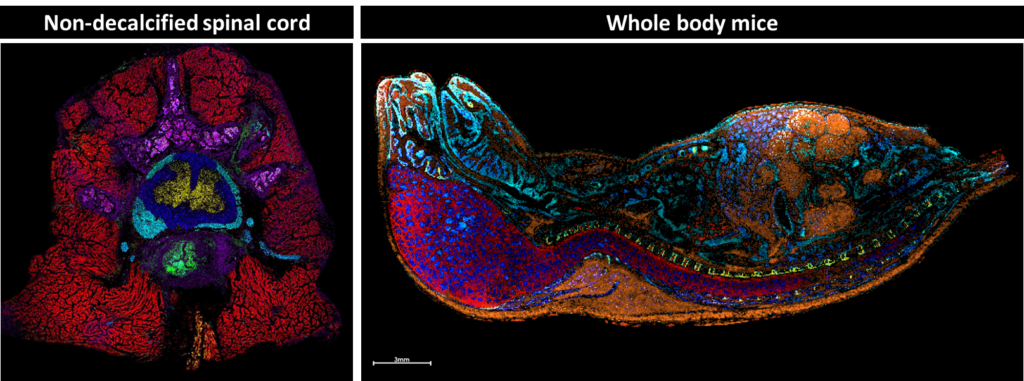
We develop methods for preparing traditionally difficult samples such as fresh-frozen, undecalcified spine and whole-body mice. This allows us to study the native metabolomics of complex and fragile samples by doing data analysis in situ, keeping the spatial context intact. These methods can be used to study development and neurological conditions while minimizing disturbances to the native molecular environment.
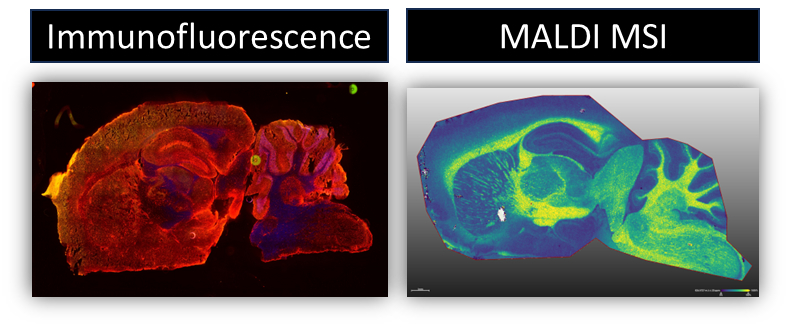
We develop methods that allow for multimodal analysis of the same sample in order to get a more wholistic view of the molecular and cellular microenvironment of samples. By combining things like MALDI MSI and Immunofluorescence microscopy on the same piece of tissue we are able to overlay cellular and molecular information to better understand disease. This can be challenging since some modalities don’t typically interact well with each other.
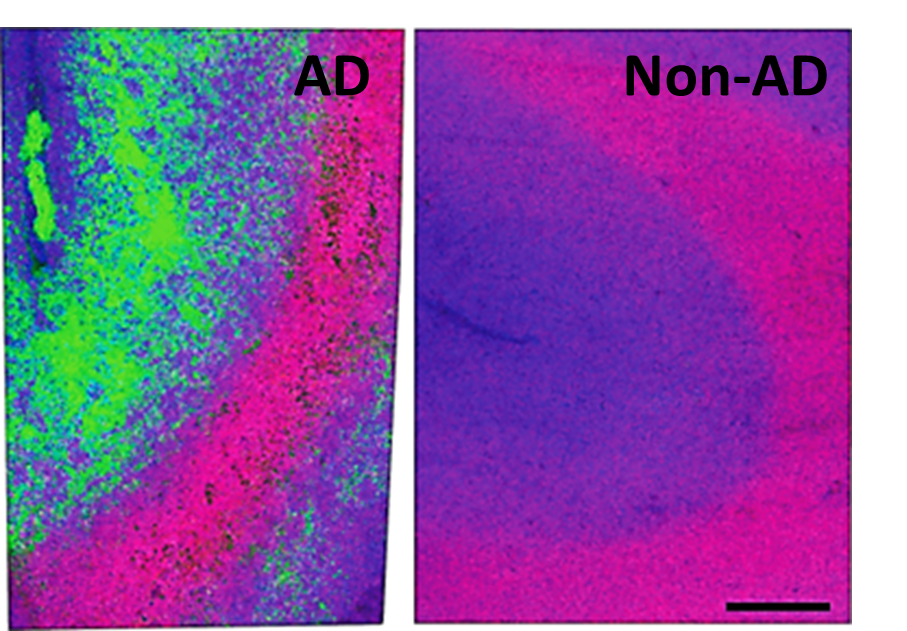
We are researching the cellular and molecular changes seen in Alzheimer’s disease (AD) as it relates to changing diet and after treatment. AD has some predictable hallmarks such as activated microglia and amyloid beta plaques, but lipidomic variation in the disease is still not clear, especially in a spatial context. We aim to show how lipids vary in the brain and how those changes relate to plaques and microglia, allowing for neighborhood analysis of the areas most effected in disease and ideally more specific therapeutic targets.